New Top-Down Proteomics Services Analyzes Intact Proteins with Post-Translationally Modified Protein
Press Release Summary:
- Ensures characterization of multiple intact proteins in a mixture
- LC, HPLC and MALDI-MS used to perform top-down proteomics studies
- Ideal for protein identification, analysis and sequence analysis
Original Press Release:
Creative Proteomics Accelerates Protein Research with Top-Down Proteomics Services
Creative Proteomics, an integrated CRO company committed to providing a full range of services in order to support various proteome-related research projects, from the identification of single proteins to large-scale proteomic studies, has recently launched its fully-developed top-down proteomics service. The service has highly supported research related to protein identification, analysis, sequence analysis and characterization of post-translational modifications.
Protein research is one of the most challenging topics in analytical chemistry. Primarily, protein research has remained focused on the development of techniques that contribute to the isolation and identification of protein sequences. The Edman degradation was a revolutionary method for amino acid sequence analysis of proteins. In the 1990s, mass spectrometry replaced Edman degradation in protein identification due to its simplicity and reliable results. Meanwhile, mass spectrometry also allowed the collection of proteins in samples. Over the years, additional protein research techniques have been developed, and the field of protein research has been defined as "proteomics". Proteomics was used in the past to define large-scale studies of the proteome, but now refers to the study of proteins from small to large scale.
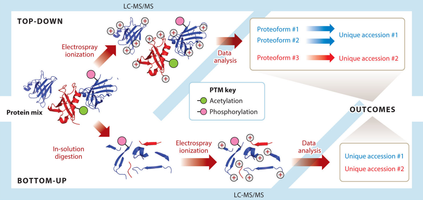
Bottom-up and top-down are two proteomics approaches based on biomass mass spectrometry. Bottom-up, also known as shotgun, is a widely used mass spectrometry technique in proteomics research. It is the traditional method of hydrolysis/enzymatic digestion for large protein fragments into peptides, which are then analyzed. On the other hand, the top-down approach directly analyzes intact proteins, including post-translationally modified proteins and other large fragments of proteins, let alone peptides.
Although several protein separation techniques have been developed, most cannot isolate proteins from complex mixtures. Top-down proteomics strategies are used to characterize multiple intact proteins in a mixture. In top-down proteomics, intact proteins are first separated from a complex biological sample through reversed-phase liquid chromatography and then directly ionized by electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI) techniques. The resulting ions are fragmented by collision-induced dissociation (CID), high-energy collision-induced dissociation (HCD), electron capture dissociation (ECD), or electron transfer dissociation (ETD) and analyzed in tandem with mass chromatography.
“We use state-of-the-art liquid chromatography (LC), high performance liquid chromatography (HPLC), and matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) to help researchers perform top-down proteomics studies, including protein sequencing, PTMs characterization, and protein structure characterization, faster, more accurately, and more efficiently,”said a senior scientist of Creative Proteomics.
About Creative Proteomics
Creative Proteomics, an experienced proteomics, metabolomics, and bioinformatics services supplier, is well equipped with advanced technologies in order to meet the increasing needs of scientists in proteomics and metabolomics-related research.
Contact:
Address: Shirley, NY 11967, USA
Email: contact@creative-proteomics.com




